Contents
- Submitting a job
- Frequently asked questions
-
- Where can I find the details about the method/algorithm?
- How do I retrieve results?
- How to create *.tar.gz file that meet the requirements?
- What does the result look like?
- Can I skip this page and directly submit the result to the backend?
- What if I have more questions/my task failed and I could not figure out why?
- How long can I access the results?
- What is Native-like Template Filter?
- Interpreting results
- Browser compatibility
- Intergrating PSICA into your own pipeline
Submitting a job
- Protein sequence field:
- Use single letter code. One plain sequence, no headers, no white space, alphabetical (A-Z, a-z) characters only.
- e.g. GHMEGKPKMEPAASSQAAVEELRTQVRELRSIIETMKDQQKREIKQLLSELDEEKKIRLRLQMEVNDIKKALQSK
- Upload Decoys (*.pdb or *.tar.gz, MaxSize:100MB):
- Use *.pdb format when only have one predicted structure, compress files to *.tar.gz when have multiple predicted structure. For more information on creating *.tat.gz files, please refer to "How to create *.tar.gz file that meet the requirements?"
- The "Residue sequence number"(column 23-26) in *.pdb files must match its position in "Protein sequence" field.
- e.g. download file in here
- Email field (Optional):
- One email address only;
- The result should be sent to the email address provided in this filed, if you did not receive the result email after a long period of time, please refer to "How do I retrieve results?"section for more information;
- Target Name field (Optional):
- Alphanumeric (A-Z, a-z, 0-9) characters only, allow 1-150 characters.
- e.g. T0865
Frequently asked questions
- Where can I find the details about the method/algorithm?
- Please refer to: W. Wang, J. Wang, D. Xu and Y. Shang, "Two New Heuristic Methods for Protein Model Quality Assessment," in IEEE/ACM Transactions on Computational Biology and Bioinformatics. doi:10.1109/TCBB.2018.2880202
- How do I retrieve results?
- Method_1. After a SUCCESSFUL submission, the browser should also ask you to download a task receipt file (e.g. [Task receipt]), open the file with text editor, it will tell you the URL to see the result (or the task status if not yet finished)
- Method_2. After a SUCCESSFUL submission, you will be automatically redirected to the task statues page, when the task is finished, the result will be shown on this page. (Note: this page does not refresh automatically, you need to do it manually)
- Method_3. After the task is finished, our server will send out the result to the email address provided, but due to the limitation of frequency our server is allowed to send out emails, and anti-spam settings on your email service provide. We notice the email might get lost in the process, so please use the above two methods to retrive the result
- Note: we will clean up the result periodically to free up the space, so please save the result to your local storage devices.
- How to create *.tar.gz file that meet the requirements?
- Please make sure none (folder,model or tarball) of the file names contains whitespace
- For Linux, MacOS, and Unix users Video Tutorial
-
- Tarball the folder directly containing the *.pdb files. Run this command OUTSIDE of the directory, NOT INSIDE!
tar cvf decoy_dir.tar decoy_dir/ - Gzip the tar file
gzip decoy_dir.tar
- Tarball the folder directly containing the *.pdb files. Run this command OUTSIDE of the directory, NOT INSIDE!
- For windows users Video Tutorial
-
- We provide tarball generator tool that can be downloaded in here (command-line interface, please run using cmd.exe or PowerShell)
- What does the result look like?
- If the task is still running [task statues]
- If the task is finished [Results-analysis for individual decoy] [Results-summary]
- Can I skip this page and directly submit the result to the backend?
- Yes, and we also provide scripts to help you integrate our server to your own pipeline.Please refer to [Intergrating PSICA into your own pipeline].
- What if I have more questions/my task failed and I could not figure out why?
- You may contact us via email: QAS_admin[anti-spam]w-wb.org(replace [anti-spam] with @)
- How long can I access the results?
- Currently we can guarantee at least 6 weeks of retention time counting from when the task is finished.
- What is Native-like Template Filter?
- It is not rare that a newly designed QA algorithm performs very well during the internal test but fails during blind test (i.e. CASP). For the template-based methods, one of the possible reasons is that there are native-like structures in the database during the internal testing. Enabling this option will remove those templates that satisfy both Coverage>94% and Indentities>94%, thus mitigate the issue with native-like structures. Generally, you do not need to enable this option during the blind test or simply want to know quality of the predicted structures.
Interpreting results
| MUfoldQA_C add-on disabled | MUfoldQA_C add-on enabled |
|
|
Click picture to view demo
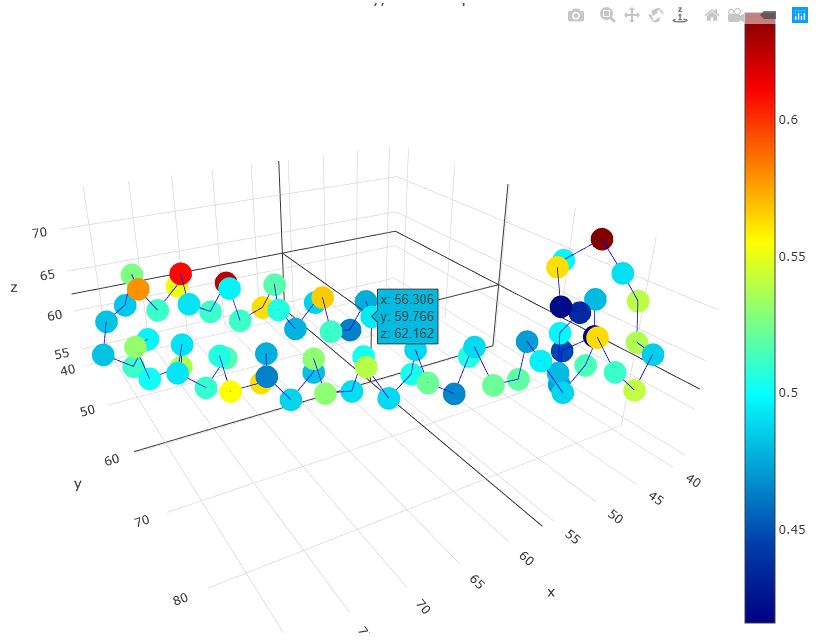
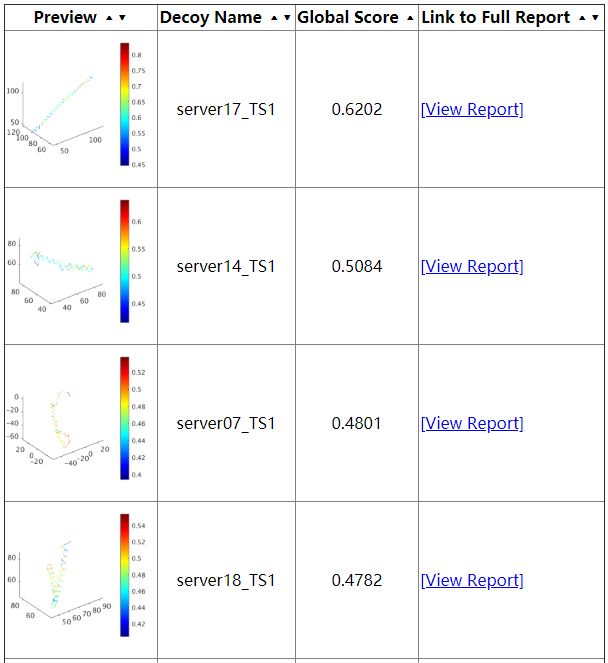
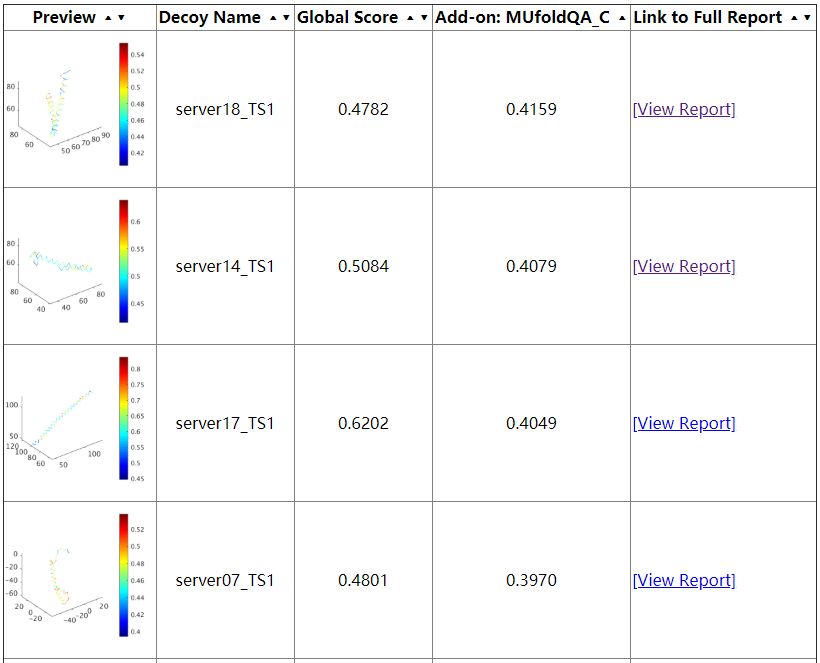
The first thing user will see is the summary page (as shown above). This page provides a table of all the decoys been evaluated as well as their corresponding global scores. This score is an estimation of GDT-TS score, in the range of 0 to 1, higher the better, usually, if the score is larger than 0.5, then the predicted model is similar to the true structure, if the score is below 0.2, it usually indicates that there are some serious misfold exist in the model. Following the global score, are the links to a more comprehensive report on each indiviosual decoys been evaluated.
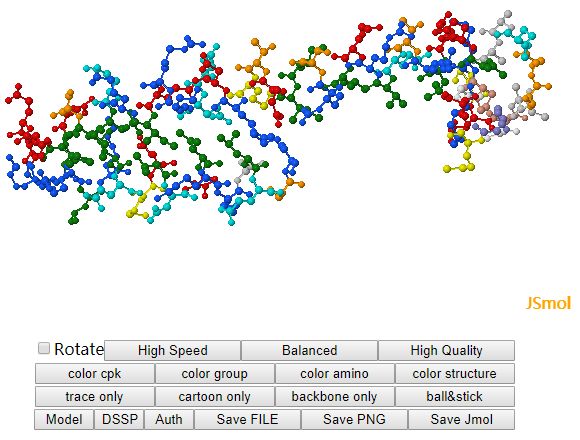
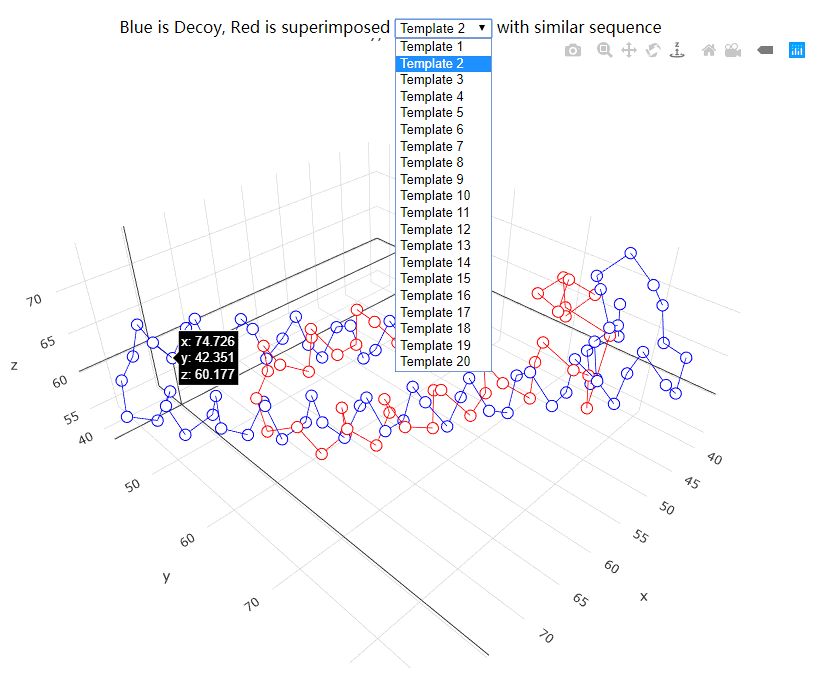
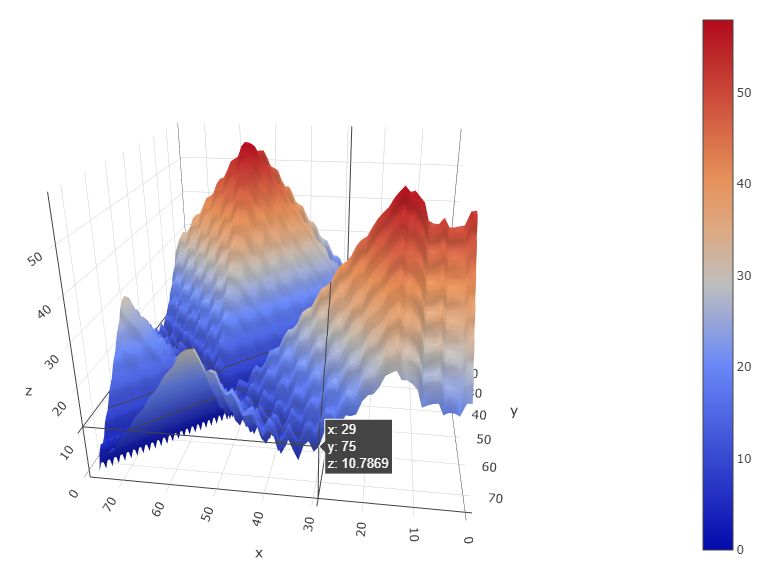
The individual decoy analysis page (as shown above) visualizes the local quality of the protein, the local score is also in the range of 0 to 1, higher the better. Combined with interactive decoy-template comparison, they can help to decide which part has low quality and how to improve it. The JSmol and DistMat visualization are also provided to provide convenient one-stop solution.
Browser compatibility
| Chrome | Firefox | Internet Explorer | Microsoft Edge | Opera | Safari | |
|---|---|---|---|---|---|---|
| Mac | Supported | Supported | N/A | N/A | Supported | Supported |
| Windows | Supported | Supported | Supported, IE10+ | Supported | Supported | N/A |
| Linux | Supported* | Distro Dependent* | N/A | N/A | Supported | N/A |
*Although everthing works as expected during our tests, there have been reports that the combination of certain version of Firefox and certain version of Linux distribution may cause Plotly© (one of the libraries we used to provide interactive user experience) not functioning as expected. Please use another browser to view such content. This does not in anyway affect the accuracy of our scores.
The above table was generated based on the following OS and browser combination
- Mac OS 10.14.4 (18E226):
- Chrome 73.0.3683.103 (Official Build) (64-bit)
- Firefox 66.0.3 (64-bit)
- Safari 12.1 (14607.1.40.1.4)
- Opera 60.0.3255.27
- Windows 10.0.17134.556:
- Chrome 73.0.3683.103 (Official Build) (64-bit)
- Firefox 66.0.3 (64-bit)
- Internet Explorer 11.523.17134.0
- Microsoft Edge 42.17134.1.0 (Microsoft EdgeHTML 17.17134)
- Opera 58.0.3135.131
- Ubuntu 16.04.3 LTS:
- Chrome 73.0.3683.103 (Official Build) (64-bit)
- Firefox 66.0.2 (64-bit)
- Opera 60.0.3255.27
Intergrating PSICA into your own pipeline
For developers who wish to intergrate PSICA to their own pipeline, we provide a fully automatic script that demonstrates the entire process of submitting a job, check job status, and parse the result when the job is done. This script mainly uses system calls to curl plus some regex, which makes it easy to port to programming languages you are comfortable with.